5 Enumeration
Polytomous LCA deals with variables that have more than two categories, such as survey questions with responses like never, sometimes, and always. The workflow of a polytomous LCA model is similar to that of an LCA model with binary indicators. However, polytomous LCA captures more complex response patterns, which can make interpretation more complex. The following code demonstrates an example, along with a visualization of the model.
5.1 Example: Elections
“Two sets of six questions with four responses each, asking respondents’ opinions of how well various traits describe presidential candidates Al Gore and George W. Bush. Also potential covariates vote choice, age, education, gender, and party ID. Source: The National Election Studies (2000).” (poLCA, 2016) See documentation here
Two sets of six questions with four responses each, asking respondents’ opinions of how well various traits describe presidential candidates Al Gore and George W. Bush. In the election data set, respondents to the 2000 American National Election Study public opinion poll were asked to evaluate how well a series of traits—moral, caring, knowledgeable, good leader, dishonest, and intelligent—described presidential candidates Al Gore and George W. Bush. Each question had four possible choices: (1) extremely well; (2) quite well; (3) not too well; and (4) not well at all.
Load packages
library(poLCA)
library(tidyverse)
library(janitor)
library(gt)
library(MplusAutomation)
library(here)
library(glue)5.2 Prepare Data
data(election)
# Detaching packages that mask the dpylr functions
detach(package:poLCA, unload = TRUE)
detach(package:MASS, unload = TRUE)
df_election <- election %>%
clean_names() %>%
select(moralb:intelb) %>%
mutate(across(everything(),
~ as.factor(as.numeric(gsub("\\D", "", .))),
.names = "{.col}1"))
# Quick summary
summary(df_election)
#> moralb caresb
#> 1 Extremely well :340 1 Extremely well :155
#> 2 Quite well :841 2 Quite well :625
#> 3 Not too well :330 3 Not too well :562
#> 4 Not well at all: 98 4 Not well at all:342
#> NA's :176 NA's :101
#> knowb leadb
#> 1 Extremely well :274 1 Extremely well :266
#> 2 Quite well :933 2 Quite well :842
#> 3 Not too well :379 3 Not too well :407
#> 4 Not well at all:133 4 Not well at all:166
#> NA's : 66 NA's :104
#> dishonb intelb moralb1
#> 1 Extremely well : 70 1 Extremely well :329 1 :340
#> 2 Quite well :288 2 Quite well :967 2 :841
#> 3 Not too well :653 3 Not too well :306 3 :330
#> 4 Not well at all:574 4 Not well at all:110 4 : 98
#> NA's :200 NA's : 73 NA's:176
#> caresb1 knowb1 leadb1 dishonb1 intelb1
#> 1 :155 1 :274 1 :266 1 : 70 1 :329
#> 2 :625 2 :933 2 :842 2 :288 2 :967
#> 3 :562 3 :379 3 :407 3 :653 3 :306
#> 4 :342 4 :133 4 :166 4 :574 4 :110
#> NA's:101 NA's: 66 NA's:104 NA's:200 NA's: 735.3 Descriptive Statistics
ds <- df_election %>%
pivot_longer(moralb1:intelb1, names_to = "variable") %>%
count(variable, value) %>% # Count occurrences of each value for each variable
group_by(variable) %>%
mutate(prop = n / sum(n)) %>%
arrange(desc(variable))
# Create the table
prop_table <- ds %>%
gt() %>%
tab_header(title = md("**Descriptive Summary**")) %>%
cols_label(
variable = "Variable",
n = md("*N*"),
prop = md("Proportion")
) %>%
fmt_number(c("n", "prop"), decimals = 2) %>% # Format both n and prop columns
cols_align(
align = "center",
columns = c(prop, n)
)
# View the table
prop_table| Descriptive Summary | ||
| value | N | Proportion |
|---|---|---|
| moralb1 | ||
| 1 | 340.00 | 0.19 |
| 2 | 841.00 | 0.47 |
| 3 | 330.00 | 0.18 |
| 4 | 98.00 | 0.05 |
| NA | 176.00 | 0.10 |
| leadb1 | ||
| 1 | 266.00 | 0.15 |
| 2 | 842.00 | 0.47 |
| 3 | 407.00 | 0.23 |
| 4 | 166.00 | 0.09 |
| NA | 104.00 | 0.06 |
| knowb1 | ||
| 1 | 274.00 | 0.15 |
| 2 | 933.00 | 0.52 |
| 3 | 379.00 | 0.21 |
| 4 | 133.00 | 0.07 |
| NA | 66.00 | 0.04 |
| intelb1 | ||
| 1 | 329.00 | 0.18 |
| 2 | 967.00 | 0.54 |
| 3 | 306.00 | 0.17 |
| 4 | 110.00 | 0.06 |
| NA | 73.00 | 0.04 |
| dishonb1 | ||
| 1 | 70.00 | 0.04 |
| 2 | 288.00 | 0.16 |
| 3 | 653.00 | 0.37 |
| 4 | 574.00 | 0.32 |
| NA | 200.00 | 0.11 |
| caresb1 | ||
| 1 | 155.00 | 0.09 |
| 2 | 625.00 | 0.35 |
| 3 | 562.00 | 0.31 |
| 4 | 342.00 | 0.19 |
| NA | 101.00 | 0.06 |
# Save as a Word doc
#gtsave(prop_table, here("figures", "prop_table.docx"))5.4 Enumeration
This code uses the mplusObject function in the MplusAutomation package.
lca_enumeration <- lapply(1:6, function(k) {
lca_enum <- mplusObject(
TITLE = glue("{k}-Class"),
VARIABLE = glue(
"categorical = moralb1-intelb1;
usevar = moralb1-intelb1;
classes = c({k}); "),
ANALYSIS =
"estimator = mlr;
type = mixture;
starts = 500 100;
processors = 10;",
OUTPUT = "sampstat residual tech11 tech14 svalues;",
usevariables = colnames(df_election),
rdata = df_election)
lca_enum_fit <- mplusModeler(lca_enum,
dataout=glue(here("poLCA", "election.dat")),
modelout=glue(here("poLCA", "c{k}_election.inp")) ,
check=TRUE, run = TRUE, hashfilename = FALSE)
})5.4.1 Table of Fit
source(here("functions","enum_table.R"))
output_election <- readModels(here("poLCA"), filefilter = "election", quiet = TRUE)
# To see rows:
#seeRows(output_election)
# Arguments for `enum_table`
# 1. readModels objects
# 2-5. Rows of successfully estimated models
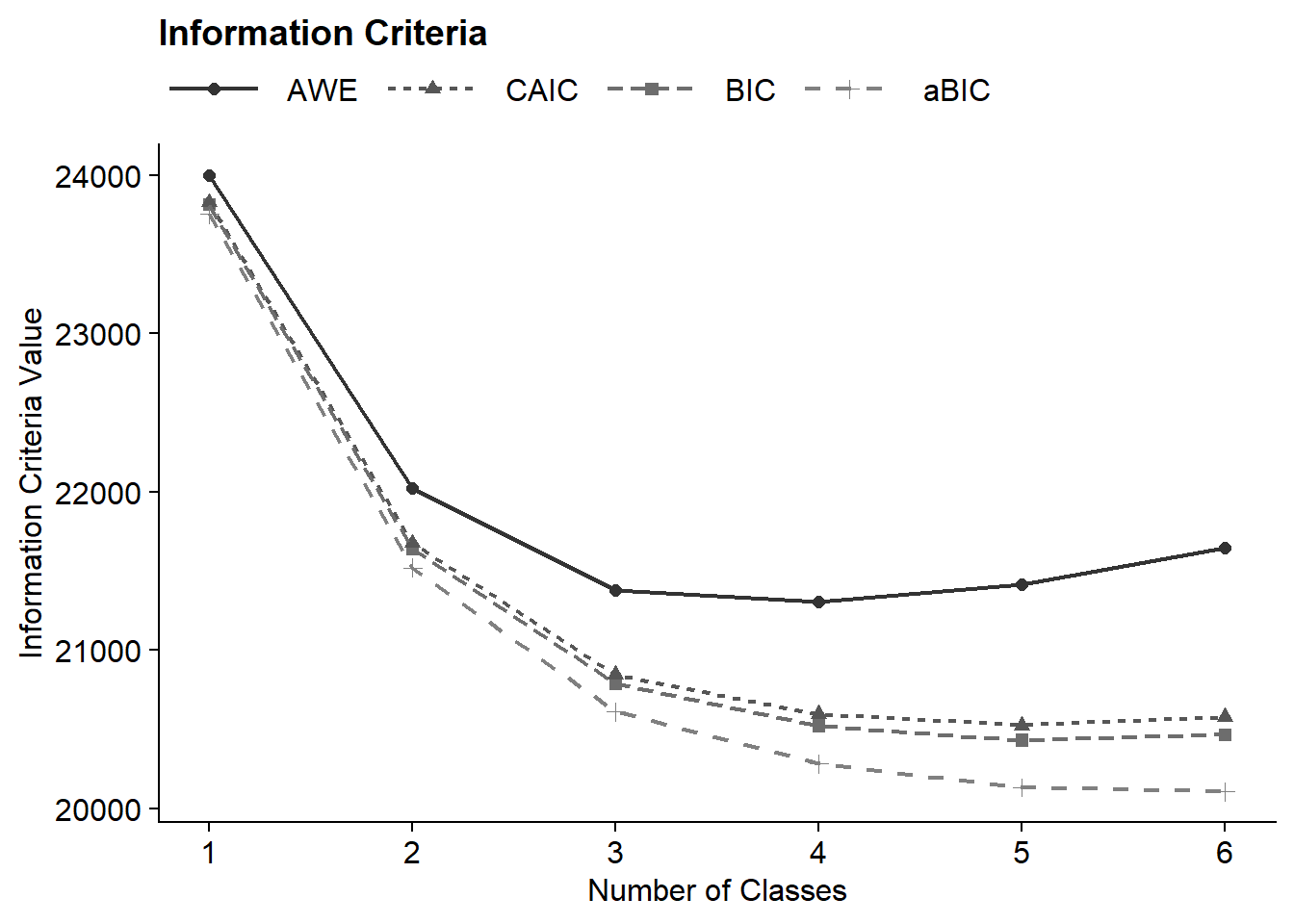
fit_table <- enum_table(output_election, 1:6)
fit_table| Model Fit Summary Table1 | ||||||||||
| Classes | Par | LL | BIC | aBIC | CAIC | AWE | BLRT | VLMR | BF | cmPk |
|---|---|---|---|---|---|---|---|---|---|---|
| 1-Class | 18 | −11,838.63 | 23,811.84 | 23,754.66 | 23,829.84 | 24,000.42 | – | – | 0.0 | <.001 |
| 2-Class | 37 | −10,680.33 | 21,637.30 | 21,519.75 | 21,674.30 | 22,024.92 | <.001 | <.001 | 0.0 | <.001 |
| 3-Class | 56 | −10,184.89 | 20,788.46 | 20,610.55 | 20,844.46 | 21,375.14 | <.001 | <.001 | 0.0 | <.001 |
| 4-Class | 75 | −9,979.58 | 20,519.89 | 20,281.62 | 20,594.89 | 21,305.63 | <.001 | <.001 | 0.0 | <.001 |
| 5-Class | 94 | −9,863.42 | 20,429.62 | 20,130.99 | 20,523.62 | 21,414.41 | <.001 | <.001 | >100 | 1.000 |
| 6-Class | 113 | −9,809.63 | 20,464.09 | 20,105.10 | 20,577.09 | 21,647.93 | <.001 | 0.629 | – | <.001 |
| 1 Note. Par = Parameters; LL = model log likelihood; BIC = Bayesian information criterion; aBIC = sample size adjusted BIC; CAIC = consistent Akaike information criterion; AWE = approximate weight of evidence criterion; BLRT = bootstrapped likelihood ratio test p-value; cmPk = approximate correct model probability. | ||||||||||
Save table: