22 Joint Occurrence
Example: Longitudinal Study of American Youth
Data source: : See documentation here
22.1 Load Packages
library(MplusAutomation)
library(tidyverse)
library(here)
library(glue)
library(gt)
library(cowplot)
library(kableExtra)
library(psych)
library(float)
library(janitor)
library(ggalluvial)
library(DiagrammeR)
library(modelsummary)
library(corrplot)
library(ggrepel)22.2 Path Diagram
| LCA Indicators: Math and Science | |
| Name | Description |
|---|---|
| Math | |
| KA46A | I Enjoy Math |
| KA46H | Math is Useful in Everyday Problems |
| KA46I | Math Helps Logical Thinking |
| KA46K | Need Math for a Good Job |
| KA46L | Will Use Math Often as an Adult |
| Science | |
| KA47A | I Enjoy Science |
| KA47H | Science is Useful in Everyday Problems |
| KA47I | Science Helps Logical Thinking |
| KA47K | Need Science for a Good Job |
| KA47L | Will Use Science Often as an Adult |
Read in LSAY dataset
data <- read_csv(here("data", "lsay_joint_occurrence.csv")) %>%
rename(
math_enjoy = KA46A, # Renaming the variables
math_useful = KA46H,
math_logical = KA46I,
math_job = KA46K,
math_adult = KA46L,
sci_enjoy = KA47A,
sci_useful = KA47H,
sci_logical = KA47I,
sci_job = KA47K,
sci_adult = KA47L
) %>%
clean_names() %>% # Making variables lower-case
mutate(across(
.cols = math_enjoy:sci_adult, # Dichtomizing the variables
.fns = ~ case_when(
. %in% c(1, 2) ~ 1,
. %in% c(3, 4, 5) ~ 0,
TRUE ~ NA_real_
)
))22.3 Descriptive Statistics
22.3.1 Descriptive Statistics using R:
Quick view of all the relevant variables:
data %>%
select(
math_enjoy, math_useful, math_logical, math_job, math_adult,
sci_enjoy, sci_useful, sci_logical, sci_job, sci_adult
) %>%
psych::describe()Proportion of indicators using R:
# Set up data to find proportions of binary indicators
ds <- data %>%
pivot_longer(c(math_enjoy:sci_adult), names_to = "Variable")
# Create table of variables and counts
tab <- table(ds$Variable, ds$value)
# Find proportions and round to 3 decimal places
prop <- prop.table(tab, margin = 1) %>%
round(3)
# Combine everything to one table
dframe <- data.frame(Variables=rownames(tab), Proportion=prop[,2], Count=tab[,2])
#remove row names
row.names(dframe) <- NULL
# Create table
gt(dframe) %>%
tab_header(title = md("**LCA Indicator Proportions**"), subtitle = md(" ")) %>%
tab_options(column_labels.font.weight = "bold", row_group.font.weight = "bold") %>%
tab_row_group(group = "Math", rows = 1:5) %>%
tab_row_group(group = "Science", rows = 6:10) %>%
row_group_order(groups = c("Math","Science")) %>%
tab_options(column_labels.font.weight = "bold", row_group.font.weight = "bold") | LCA Indicator Proportions | ||
| Variables | Proportion | Count |
|---|---|---|
| Math | ||
| math_adult | 0.634 | 2121 |
| math_enjoy | 0.547 | 1847 |
| math_job | 0.567 | 1891 |
| math_logical | 0.680 | 2279 |
| math_useful | 0.635 | 2131 |
| Science | ||
| sci_adult | 0.403 | 1352 |
| sci_enjoy | 0.534 | 1793 |
| sci_job | 0.341 | 1139 |
| sci_logical | 0.548 | 1825 |
| sci_useful | 0.450 | 1502 |
Data summary:
select_data <- data %>%
select(math_enjoy:sci_adult)
f <- All(select_data) ~ Mean + SD + Min + Median + Max + Histogram
datasummary(f, data, output="markdown")| Mean | SD | Min | Median | Max | Histogram | |
|---|---|---|---|---|---|---|
| math_enjoy | 0.55 | 0.50 | 0.00 | 1.00 | 1.00 | ▆▇ |
| math_useful | 0.63 | 0.48 | 0.00 | 1.00 | 1.00 | ▄▇ |
| math_logical | 0.68 | 0.47 | 0.00 | 1.00 | 1.00 | ▃▇ |
| math_job | 0.57 | 0.50 | 0.00 | 1.00 | 1.00 | ▅▇ |
| math_adult | 0.63 | 0.48 | 0.00 | 1.00 | 1.00 | ▄▇ |
| sci_enjoy | 0.53 | 0.50 | 0.00 | 1.00 | 1.00 | ▆▇ |
| sci_useful | 0.45 | 0.50 | 0.00 | 0.00 | 1.00 | ▇▆ |
| sci_logical | 0.55 | 0.50 | 0.00 | 1.00 | 1.00 | ▆▇ |
| sci_job | 0.34 | 0.47 | 0.00 | 0.00 | 1.00 | ▇▄ |
| sci_adult | 0.40 | 0.49 | 0.00 | 0.00 | 1.00 | ▇▅ |
Correlation table:
select_data %>%
datasummary_correlation(output = "markdown")| math_enjoy | math_useful | math_logical | math_job | math_adult | sci_enjoy | sci_useful | sci_logical | sci_job | sci_adult | |
|---|---|---|---|---|---|---|---|---|---|---|
| math_enjoy | 1 | . | . | . | . | . | . | . | . | . |
| math_useful | .31 | 1 | . | . | . | . | . | . | . | . |
| math_logical | .32 | .51 | 1 | . | . | . | . | . | . | . |
| math_job | .31 | .43 | .38 | 1 | . | . | . | . | . | . |
| math_adult | .36 | .55 | .41 | .53 | 1 | . | . | . | . | . |
| sci_enjoy | .23 | .23 | .25 | .22 | .24 | 1 | . | . | . | . |
| sci_useful | .21 | .41 | .36 | .31 | .33 | .43 | 1 | . | . | . |
| sci_logical | .21 | .33 | .48 | .32 | .30 | .45 | .58 | 1 | . | . |
| sci_job | .22 | .26 | .27 | .42 | .29 | .37 | .50 | .43 | 1 | . |
| sci_adult | .22 | .30 | .28 | .29 | .37 | .48 | .54 | .47 | .62 | 1 |
Correlation plot:
f_cor <- data %>%
select(math_enjoy:sci_adult) %>%
cor(use = "pairwise.complete.obs")
corrplot(f_cor,
method = "circle",
type = "upper",
tl.col="black",
tl.srt=45)
22.3.2 Descriptive Statistics using MplusAutomation:
basic_mplus <- mplusObject(
TITLE = "Descriptive Statistics;",
VARIABLE =
"usevar = math_enjoy-sci_adult;
categorical = math_enjoy-sci_adult;",
ANALYSIS = "TYPE=basic;",
OUTPUT = "sampstat;",
usevariables = colnames(data),
rdata = data)
basic_mplus_fit <- mplusModeler(basic_mplus,
dataout = here("joint_occurrence", "data.dat"),
modelout = here("joint_occurrence","basic.inp"),
check = TRUE, run = TRUE, hashfilename = FALSE)View output (which is goes more into detail) or a see a brief view of descriptive statistics using get_sampstat():
# Using MplusAutomation
MplusAutomation::get_sampstat(basic_mplus_fit)
# Using base R
summary(data)22.4 Enumeration (Math Only)
This code uses the mplusObject function in the MplusAutomation package and saves all model runs in the mplus_enum folder.
lca_enum_6 <- lapply(1:6, function(k) {
lca_enum <- mplusObject(
TITLE = glue("Math Attitudes: {k}-Class"),
VARIABLE = glue(
"categorical = math_enjoy, math_useful, math_logical, math_job, math_adult;
usevar = math_enjoy, math_useful, math_logical, math_job, math_adult;
classes = c({k});"),
ANALYSIS =
"estimator = mlr;
type = mixture;
processors = 12;
starts = 500 100;",
OUTPUT = "sampstat residual tech11 tech14;",
usevariables = colnames(data),
rdata = data)
lca_enum_fit <- mplusModeler(lca_enum,
dataout=glue(here("joint_occurrence","enum_math", "data.dat")),
modelout=glue(here("joint_occurrence","enum_math", "c{k}_math.inp")) ,
check=TRUE, run = TRUE, hashfilename = FALSE)
})IMPORTANT: Before moving forward, make sure to examine each output document to ensure models were estimated normally. In this example, the last model (6-class models) did not produce reliable output and was excluded.
22.5 Enumeration (Science Only)
This code uses the mplusObject function in the MplusAutomation package and saves all model runs in the mplus_enum folder.
lca_enum_6 <- lapply(1:6, function(k) {
lca_enum <- mplusObject(
TITLE = glue("Science Attitudes: {k}-Class"),
VARIABLE = glue(
"categorical = sci_enjoy, sci_useful, sci_logical, sci_job, sci_adult;
usevar = sci_enjoy, sci_useful, sci_logical, sci_job, sci_adult;
classes = c({k});"),
ANALYSIS =
"estimator = mlr;
type = mixture;
processors = 12;
starts = 500 100;",
OUTPUT = "sampstat residual tech11 tech14;",
usevariables = colnames(data),
rdata = data)
lca_enum_fit <- mplusModeler(lca_enum,
dataout=glue(here("joint_occurrence","enum_sci", "data.dat")),
modelout=glue(here("joint_occurrence","enum_sci", "c{k}_sci.inp")) ,
check=TRUE, run = TRUE, hashfilename = FALSE)
})IMPORTANT: Before moving forward, make sure to examine each output document to ensure models were estimated normally. In this example, the last model (6-class models) did not produce reliable output and was excluded.
22.5.0.1 Fit Table
source(here("functions", "enum_table_jo.R"))
# Read model outputs
output_enum_c1 <- readModels(here("joint_occurrence", "enum_math"), quiet = TRUE)
output_enum_c2 <- readModels(here("joint_occurrence", "enum_sci"), quiet = TRUE)
# Define rows for row groups (assuming 6 models per time)
rows_m1 <- 1:6
rows_m2 <- 7:12
fit_table_jo <- fit_table_jo(output_enum_c1, output_enum_c2, rows_m1, rows_m2)
fit_table_jo| Model Fit Summary Table1 | ||||||||||
| Classes | Par | LL | BIC | aBIC | CAIC | AWE | BLRT | VLMR | BF | cmP_k |
|---|---|---|---|---|---|---|---|---|---|---|
| LCA 1 | ||||||||||
| Math Attitudes: 1-Class | 5 | −11,112.23 | 22,265.10 | 22,249.22 | 22,270.10 | 22,320.74 | – | – | 0.0 | <.001 |
| Math Attitudes: 2-Class | 11 | −9,368.02 | 18,825.46 | 18,790.50 | 18,836.45 | 18,947.86 | <.001 | <.001 | 0.0 | <.001 |
| Math Attitudes: 3-Class | 17 | −9,243.39 | 18,624.95 | 18,570.93 | 18,641.95 | 18,814.12 | <.001 | <.001 | 0.0 | <.001 |
| Math Attitudes: 4-Class | 23 | −9,204.49 | 18,595.93 | 18,522.84 | 18,618.93 | 18,851.86 | <.001 | 0.10 | >100 | 1.00 |
| Math Attitudes: 5-Class | 29 | −9,197.51 | 18,630.72 | 18,538.57 | 18,659.72 | 18,953.42 | 0.04 | 0.00 | >100 | <.001 |
| Math Attitudes: 6-Class | 35 | −9,197.04 | 18,678.55 | 18,567.34 | 18,713.55 | 19,068.02 | 1.00 | 0.48 | – | <.001 |
| LCA 2 | ||||||||||
| Science Attitudes: 1-Class | 5 | −11,315.87 | 22,672.34 | 22,656.45 | 22,677.34 | 22,727.94 | – | – | 0.0 | <.001 |
| Science Attitudes: 2-Class | 11 | −9,009.08 | 18,107.48 | 18,072.53 | 18,118.48 | 18,229.81 | <.001 | <.001 | 0.0 | <.001 |
| Science Attitudes: 3-Class | 17 | −8,814.56 | 17,767.18 | 17,713.17 | 17,784.18 | 17,956.24 | <.001 | <.001 | 0.0 | <.001 |
| Science Attitudes: 4-Class | 23 | −8,742.24 | 17,671.26 | 17,598.17 | 17,694.26 | 17,927.04 | <.001 | <.001 | >100 | 1.00 |
| Science Attitudes: 5-Class | 29 | −8,734.82 | 17,705.15 | 17,613.01 | 17,734.15 | 18,027.66 | <.001 | 0.01 | >100 | <.001 |
| Science Attitudes: 6-Class | 35 | −8,732.97 | 17,750.16 | 17,638.95 | 17,785.17 | 18,139.40 | 1.00 | 0.48 | – | <.001 |
| 1 Note. Par = Parameters; LL = model log likelihood; BIC = Bayesian information criterion; aBIC = sample size adjusted BIC; CAIC = consistent Akaike information criterion; AWE = approximate weight of evidence criterion; BLRT = bootstrapped likelihood ratio test p-value; VLMR = Vuong-Lo-Mendell-Rubin adjusted likelihood ratio test p-value; cmPk = approximate correct model probability. | ||||||||||
Save table:
22.5.1 Information Criteria Plot
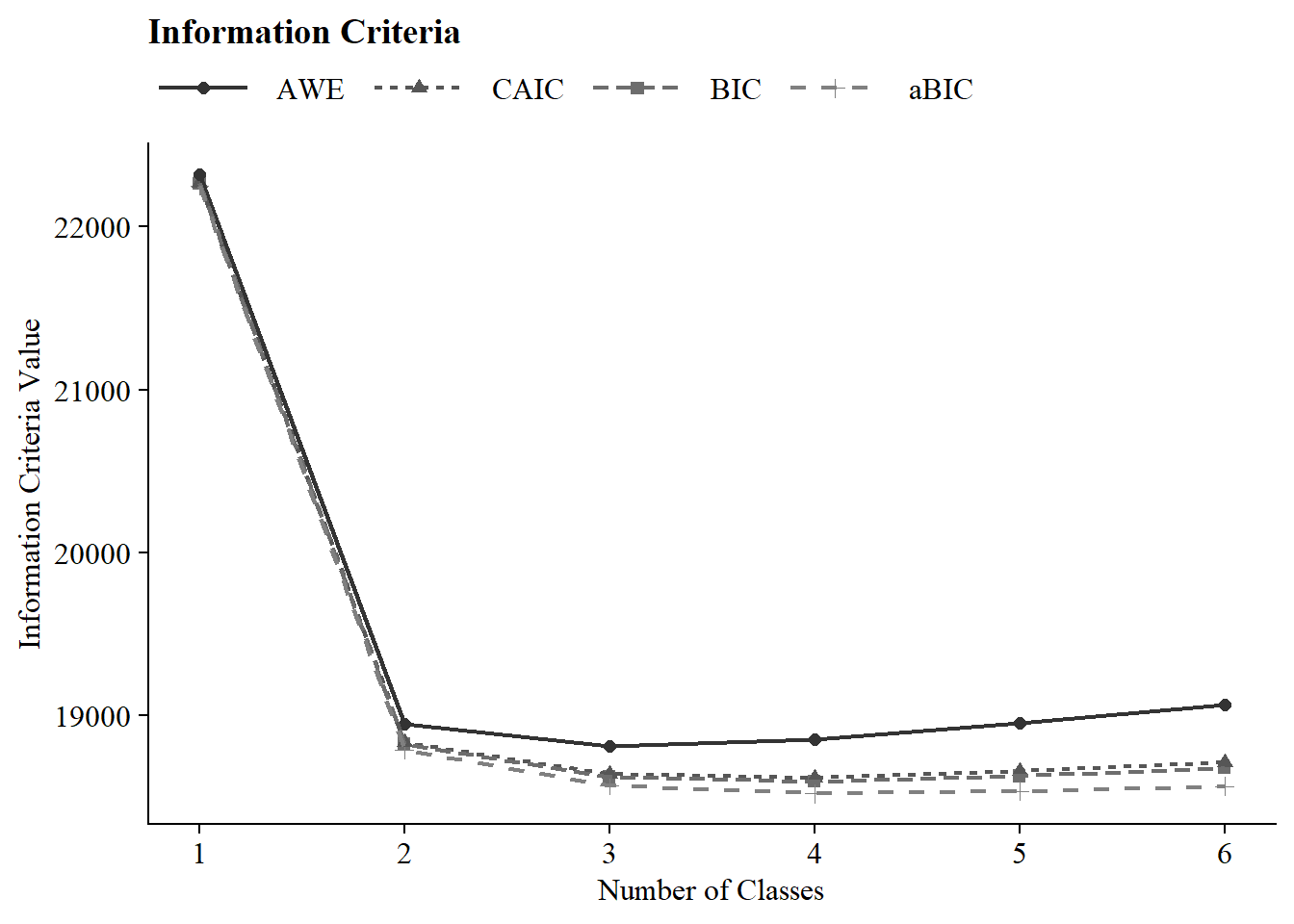
#ggsave(here("figures", "info_criteria_jo1.png"), dpi = "retina", bg = "white", height=5, width=7, units="in")
ic_plot(output_enum_c2)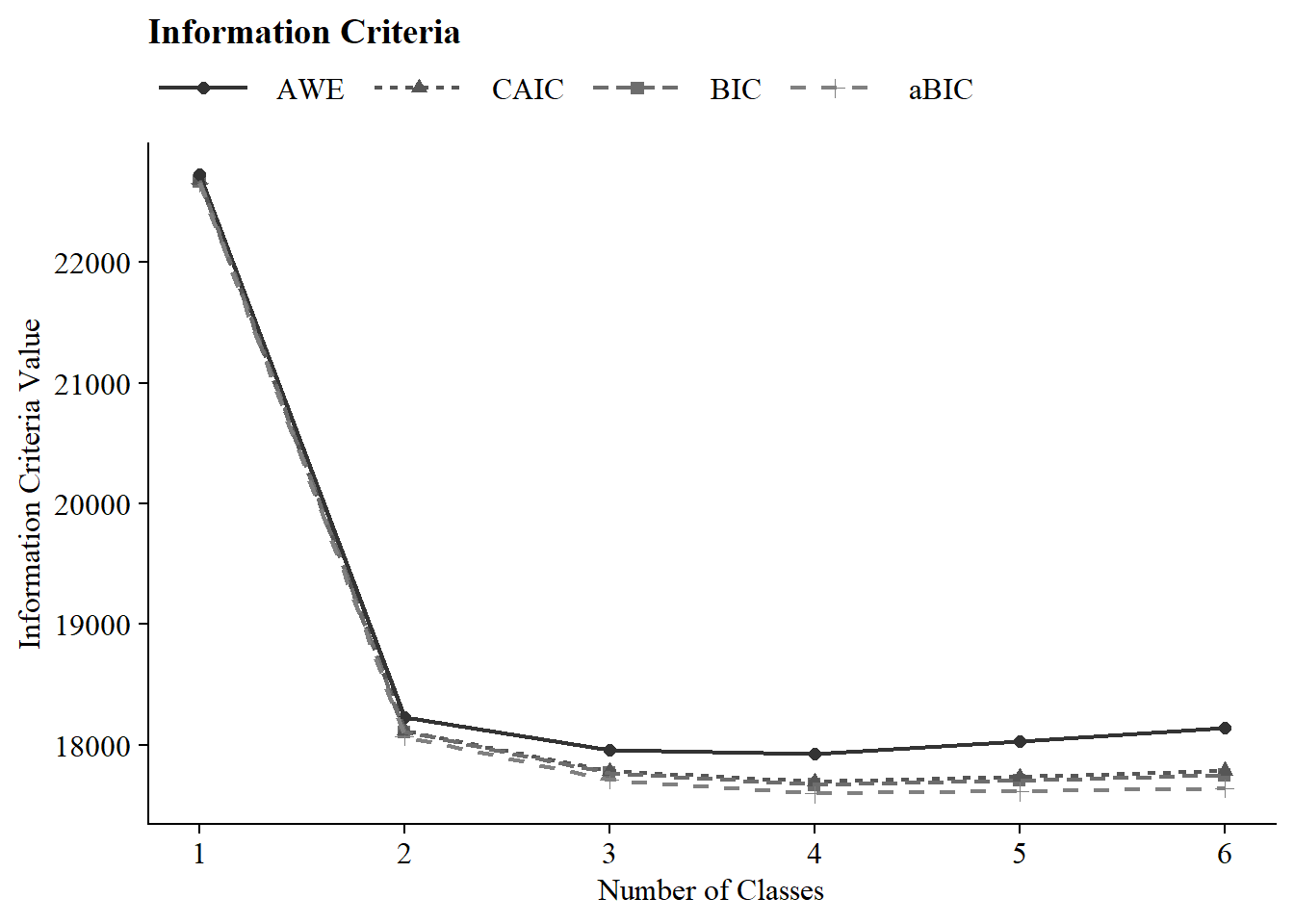
#ggsave(here("figures", "info_criteria_jo2.png"), dpi = "retina", bg = "white", height=5, width=7, units="in")22.5.2 4-Class Probability Plot
Use the plot_lca function provided in the folder to plot the item probability plot. This function requires one argument:
- model_name: The name of the Mplus readModels object (e.g., output_enum_c1$c4_math.out)
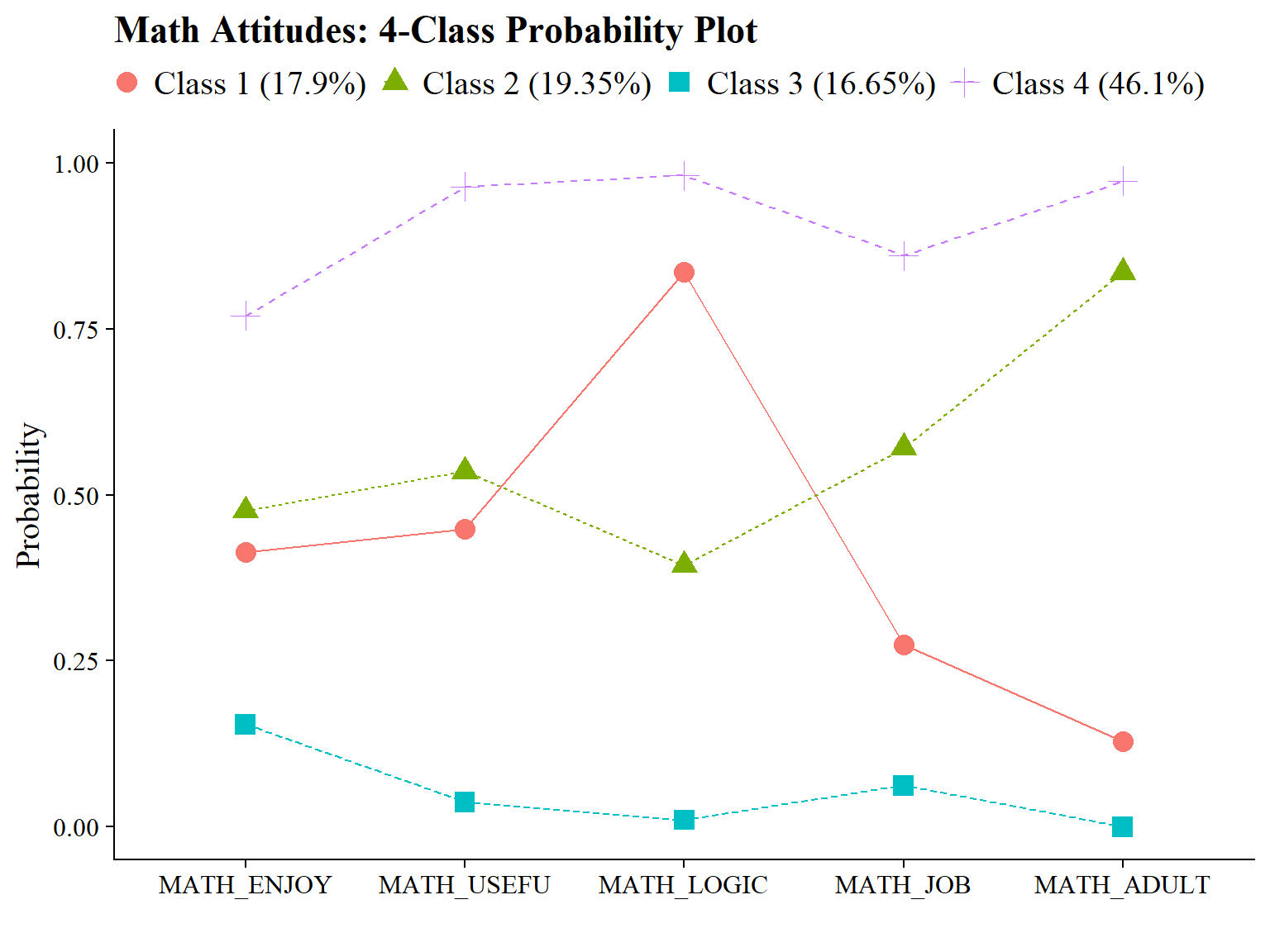
#ggsave(here("figures", "probability_plot_jo1.png"), dpi = "retina", bg = "white", height=5, width=7, units="in")
plot_lca(model_name = output_enum_c2$c4_sci.out)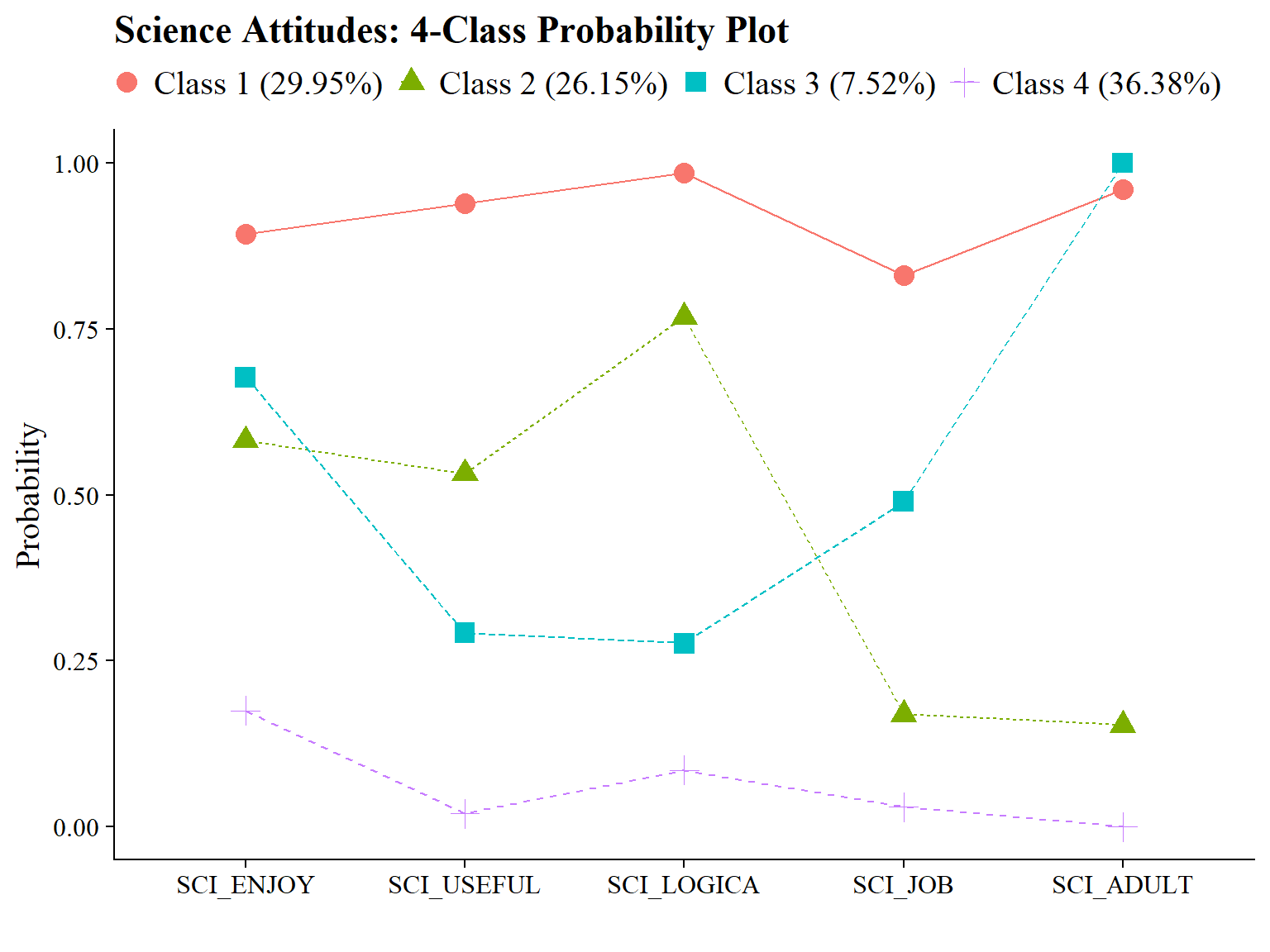
#ggsave(here("figures", "probability_plot_jo2.png"), dpi = "retina", bg = "white", height=5, width=7, units="in")22.6 Estimate Joint Occurrence LCA
22.6.1 Step 1 - Estimate Unconditional Model
Math Attitudes
Here, I included the ID variable (casenum) so I can later join the two datasets we get from step 2.
step1 <- mplusObject(
TITLE = "Step 1 - Unconditional Model",
VARIABLE = "categorical = math_enjoy, math_useful, math_logical, math_job, math_adult;
usevar = math_enjoy, math_useful, math_logical, math_job, math_adult;
idvariable = casenum;
classes = c(4);",
ANALYSIS =
"estimator = mlr;
type = mixture;
starts = 0;
OPTSEED = 830570;",
SAVEDATA =
"File=savedata_math.dat;
Save=cprob;",
OUTPUT = "sampstat residual tech11 tech14 svalues(4 1 2 3)", # I used `svalues` to rearrange the class labels
usevariables = colnames(data),
rdata = data)
step1_fit <- mplusModeler(step1,
dataout=here("joint_occurrence", "jo_model", "data.dat"),
modelout=here("joint_occurrence", "jo_model", "one_math.inp") ,
check=TRUE, run = TRUE, hashfilename = FALSE)Note: Since the emerging classes are similar between math and science, I rearranged the classes so that they match using svaues option in the OUTPUT command. For example, Class 1 of Science LCA and Class 4 of Math LCA are both the “High” class. So I changed the Math class from Class 4 to Class 1.
Evaluate output and compare the class counts and proportions for the latent classes. Using the OPTSEED function ensures replication of the best loglikelihood value run.
Science Attitudes
step1 <- mplusObject(
TITLE = "Step 1 - Unconditional Model",
VARIABLE = "categorical = sci_enjoy, sci_useful, sci_logical, sci_job, sci_adult;
usevar = sci_enjoy, sci_useful, sci_logical, sci_job, sci_adult;
idvariable = casenum;
classes = c(4);",
ANALYSIS =
"estimator = mlr;
type = mixture;
starts = 0;
OPTSEED = 761633;",
SAVEDATA =
"File=savedata_sci.dat;
Save=cprob;",
OUTPUT = "sampstat residual tech11 tech1;",
usevariables = colnames(data),
rdata = data)
step1_fit <- mplusModeler(step1,
dataout=here("joint_occurrence", "jo_model", "data.dat"),
modelout=here("joint_occurrence", "jo_model", "one_sci.inp") ,
check=TRUE, run = TRUE, hashfilename = FALSE)Confirm that plots look as expected (i.e., identical to enumeration model)
source(here("functions","plot_lca.R"))
output_math <- readModels(here("joint_occurrence","jo_model","one_math.out"))
output_sci <- readModels(here("joint_occurrence","jo_model","one_sci.out"))
plot_lca(model_name = output_math)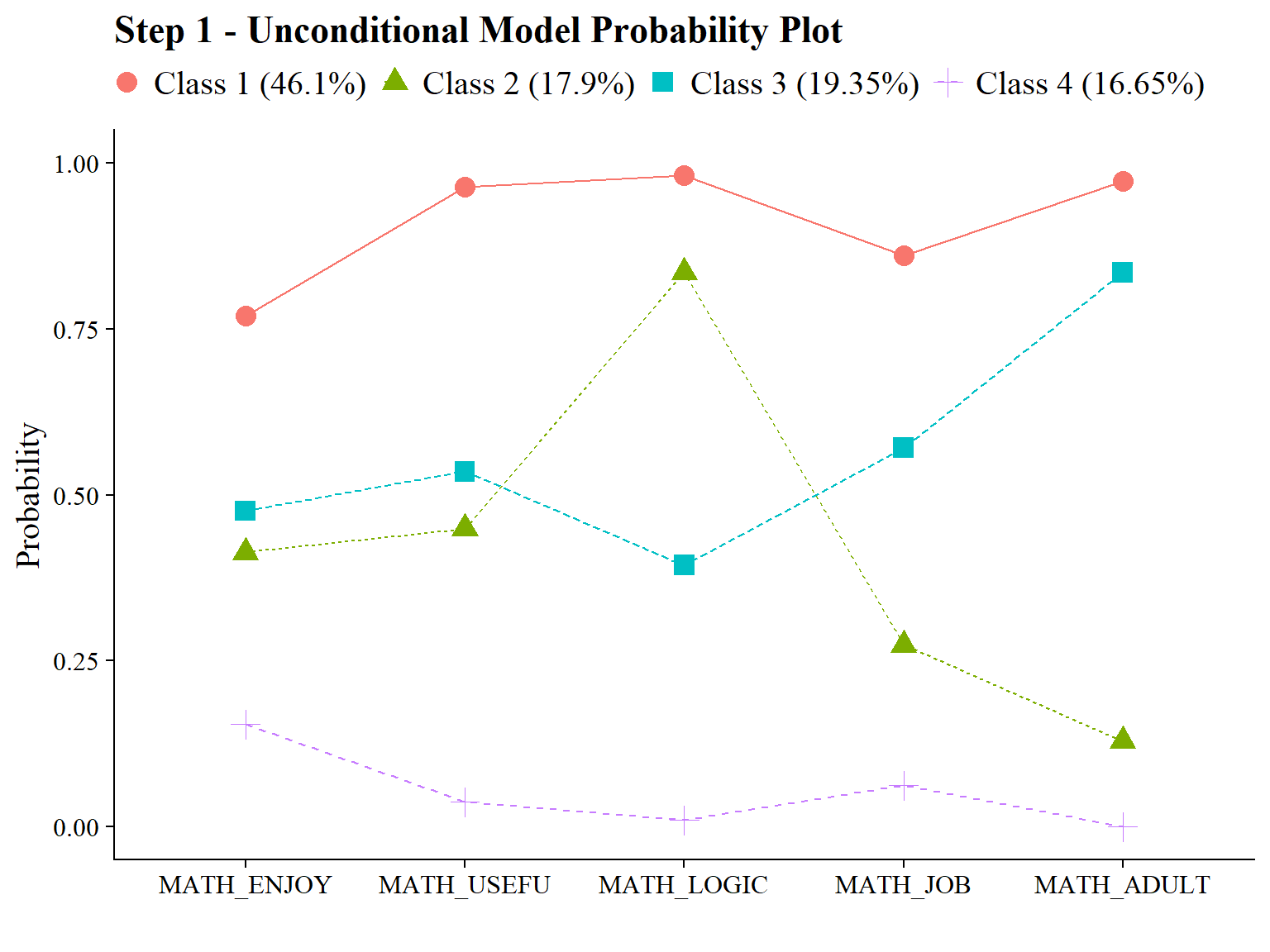
plot_lca(model_name = output_sci)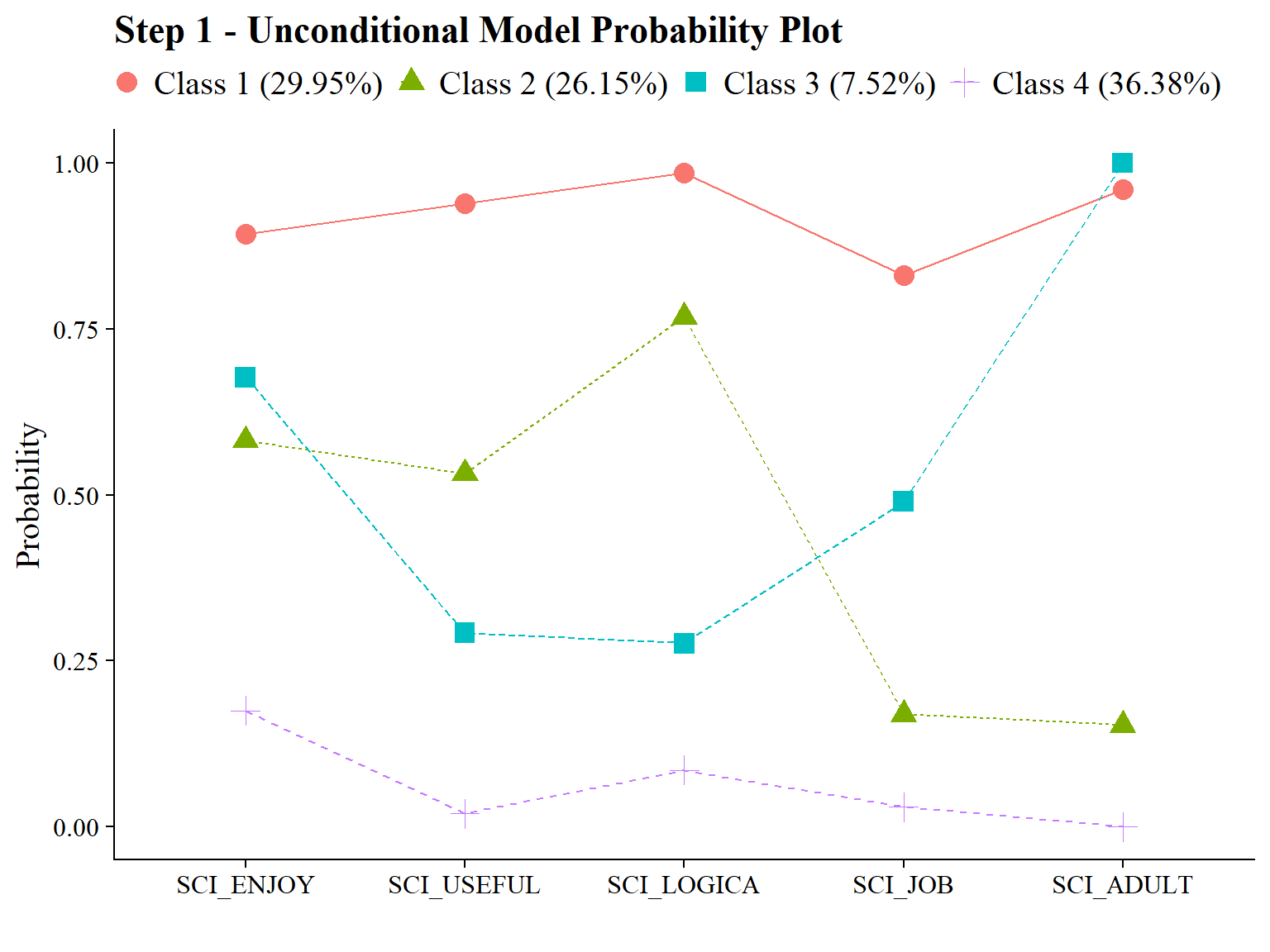
22.6.2 Step 2 - Determine Measurement Error
Extract logits for the classification probabilities for the most likely latent class:
logit_cprobs_math <- as.data.frame(output_math[["class_counts"]]
[["logitProbs.mostLikely"]])
logit_cprobs_sci <- as.data.frame(output_sci[["class_counts"]]
[["logitProbs.mostLikely"]])Extract saved dataset:
savedata_math <- as.data.frame(output_math[["savedata"]])
savedata_sci <- as.data.frame(output_sci[["savedata"]])Rename the column in savedata named “C” and change to “N”
colnames(savedata_math)[colnames(savedata_math)=="C"] <- "N_math"
colnames(savedata_sci)[colnames(savedata_sci)=="C"] <- "N_sci"
savedata <- savedata_math %>%
full_join(savedata_sci, by = "CASENUM")22.6.3 Step 3 - Add Auxiliary Variables
Build the joint occurrence model:
step3_jo <- mplusObject(
TITLE = "Joint Occurrence LCA",
VARIABLE =
"nominal=N_math N_sci;
usevar = N_math N_sci;
classes = math(4) sci(4);" ,
ANALYSIS =
"estimator = mlr;
type = mixture;
starts = 0;",
MODEL =
glue(
" %OVERALL%
sci on math;
MODEL math:
%math#1%
[N_math#1@{logit_cprobs_math[1,1]}];
[N_math#2@{logit_cprobs_math[1,2]}];
[N_math#3@{logit_cprobs_math[1,3]}];
%math#2%
[N_math#1@{logit_cprobs_math[2,1]}];
[N_math#2@{logit_cprobs_math[2,2]}];
[N_math#3@{logit_cprobs_math[2,3]}];
%math#3%
[N_math#1@{logit_cprobs_math[3,1]}];
[N_math#2@{logit_cprobs_math[3,2]}];
[N_math#3@{logit_cprobs_math[3,3]}];
%math#4%
[N_math#1@{logit_cprobs_math[4,1]}];
[N_math#2@{logit_cprobs_math[4,2]}];
[N_math#3@{logit_cprobs_math[4,3]}];
MODEL sci:
%sci#1%
[N_sci#1@{logit_cprobs_sci[1,1]}];
[N_sci#2@{logit_cprobs_sci[1,2]}];
[N_sci#3@{logit_cprobs_sci[1,3]}];
%sci#2%
[N_sci#1@{logit_cprobs_sci[2,1]}];
[N_sci#2@{logit_cprobs_sci[2,2]}];
[N_sci#3@{logit_cprobs_sci[2,3]}];
%sci#3%
[N_sci#1@{logit_cprobs_sci[3,1]}];
[N_sci#2@{logit_cprobs_sci[3,2]}];
[N_sci#3@{logit_cprobs_sci[3,3]}];
%sci#4%
[N_sci#1@{logit_cprobs_sci[4,1]}];
[N_sci#2@{logit_cprobs_sci[4,2]}];
[N_sci#3@{logit_cprobs_sci[4,3]}];"),
usevariables = colnames(savedata),
rdata = savedata)
step3_jo_fit <- mplusModeler(step3_jo,
dataout=here("joint_occurrence","jo_model","three.dat"),
modelout=here("joint_occurrence","jo_model","three.inp"),
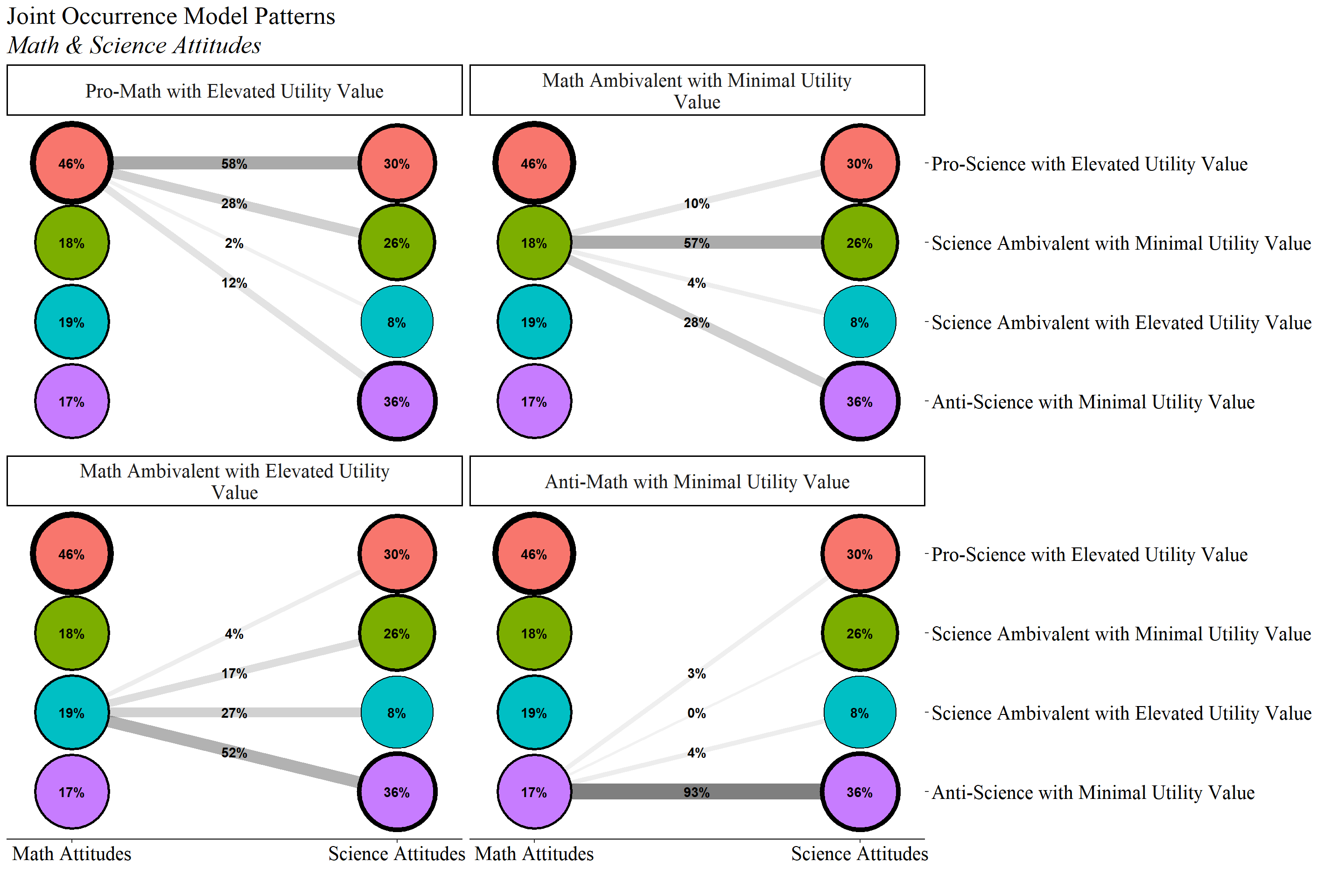
check=TRUE, run = TRUE, hashfilename = FALSE)22.6.3.1 Joint Distribution
Plot:
jo_output <- readModels(here("joint_occurrence","jo_model","three.out"))
plot_lca(model_name = output_math)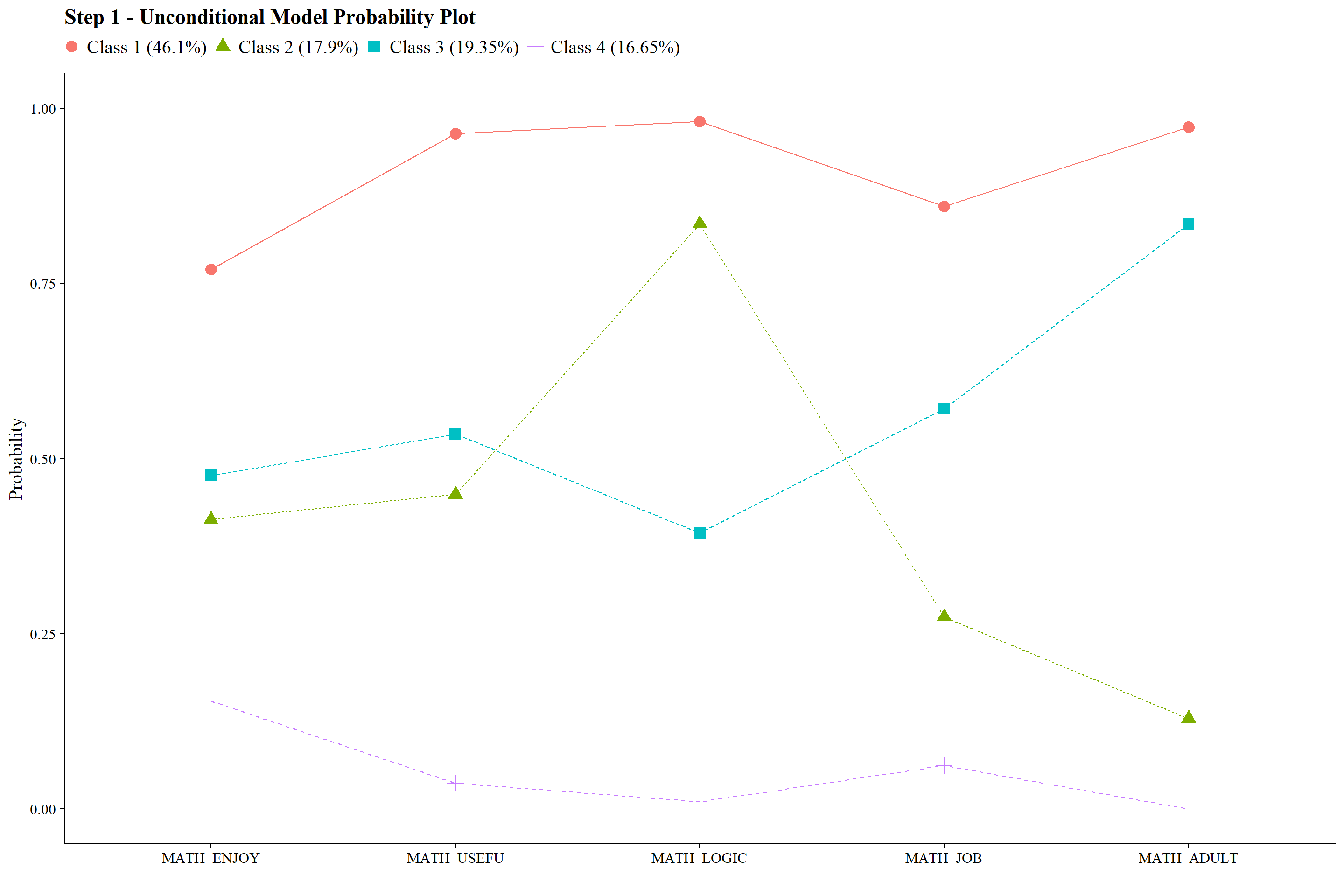
plot_lca(model_name = output_sci)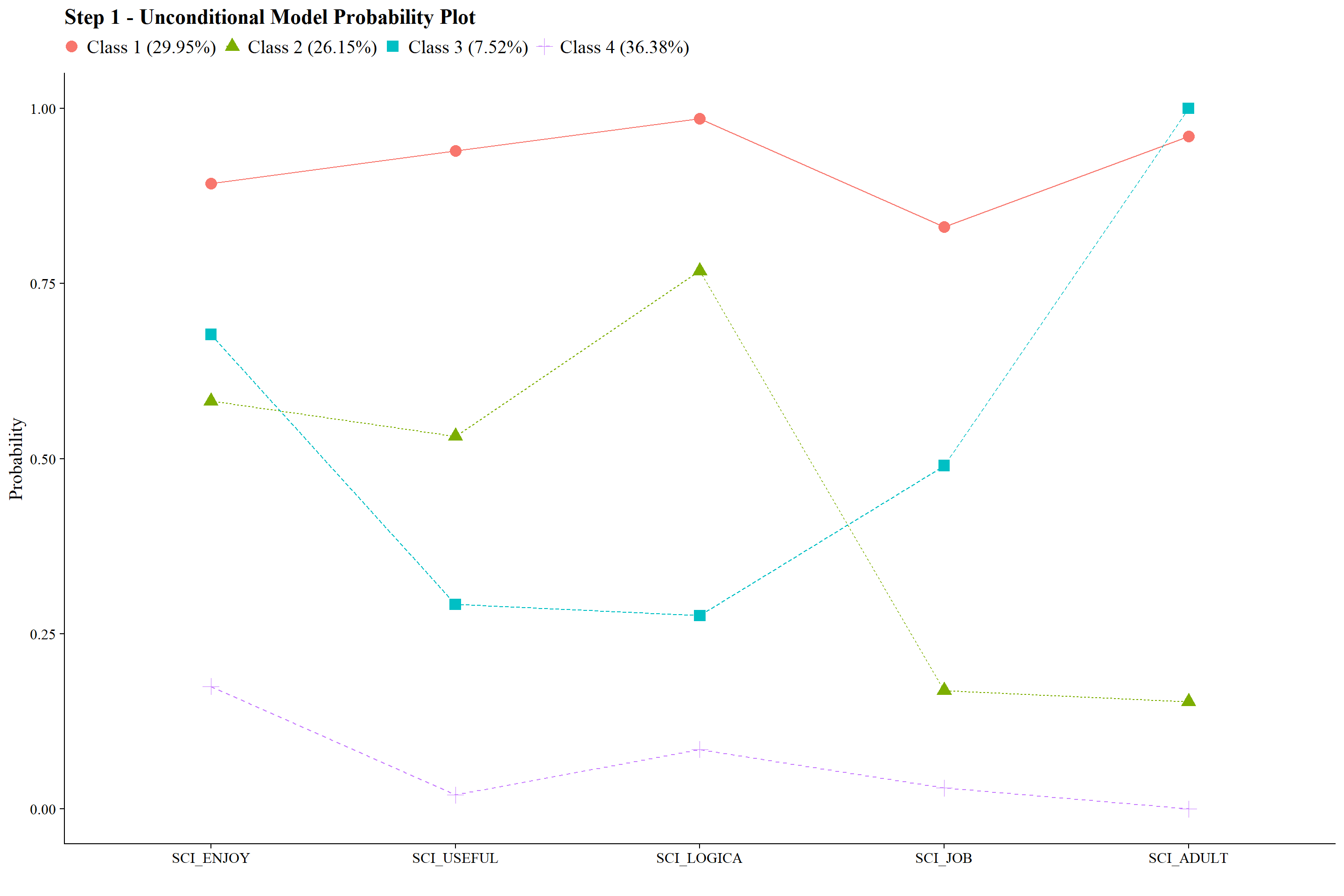
source(here("functions", "plot_patterns.R"))
title <- "Joint Occurrence Model Patterns"
subtitle <- "Math & Science Attitudes"
plot_patterns(
model_name = jo_output,
facet_labels =c( # These are the Math labels
`1` = "Pro-Math with Elevated Utility Value",
`2` = "Math Ambivalent with Minimal Utility Value",
`3` = "Math Ambivalent with Elevated Utility Value",
`4` = "Anti-Math with Minimal Utility Value"),
lca_labels = c('1' = "Math Attitudes", '2' = "Science Attitudes"),
class_labels = c( # These are the Science labels
"Pro-Science with Elevated Utility Value",
"Science Ambivalent with Minimal Utility Value",
"Science Ambivalent with Elevated Utility Value",
"Anti-Science with Minimal Utility Value"
),
title,
subtitle
)
#ggsave(here("figures","interdependencies_plot.png"), dpi=500,bg = "white", height=7, width=12, units="in")Alternative plot:
jo_output <- readModels(here("joint_occurrence","jo_model","three.out"))
jo_prob <- as.data.frame(jo_output$class_counts$transitionProbs$probability)
c1_labels <- c("Pro-Math \nwith Elevated Utility Value \n(46%)",
"Math Ambivalent \nwith Minimal Utility Value\n(18%)",
"Math Ambivalent \nwith Elevated Utility Value\n(19%)",
"Anti-Math \nwith Minimal Utility Value\n(17%)")
c2_labels <- c("Pro-Science \nwith Elevated Utility Value\n(30%)",
"Science Ambivalent \nwith Minimal Utility Value\n(26%)",
"Science Ambivalent \nwith Elevated Utility Value\n(8%)",
"Anti-Science \nwith Minimal Utility Value\n(36%)")
# T1 → T2
c1_c2 <- expand.grid(C1 = c1_labels, C2 = c2_labels) %>%
mutate(P12 = jo_prob[1:nrow(jo_prob), 1]) %>%
mutate(P12 = round(P12, 2))
# Plot for T1 -> T2
ggplot(c1_c2, aes(axis1 = C1, axis2 = C2, y = P12)) +
geom_alluvium(aes(fill = C1), width = 0.2, alpha = 0.7) +
# Make the stratum rectangles white instead of gray:
geom_stratum(width = 0.2, color = "black") +
geom_text(
stat = "stratum",
aes(label = after_stat(stratum)),
size = 3.5
) +
# Label the flows themselves with the probability
# geom_text(aes(label = P12),
# stat = "flow", nudge_x = .2, size = 5) +
scale_x_discrete(limits = c("Math Attitudes", "Science Attitudes"), expand = c(.1, .1)) +
labs(subtitle = "Math and Science Attitudes", title = "Joint Occurrence Model", x = "") +
theme_minimal() +
theme(
text = element_text(family = "serif", size = 20),
legend.position = "none",
axis.text.x = element_text(color = "black"),
axis.title.y = element_blank(),
axis.text.y = element_blank(),
axis.ticks.y = element_blank(),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
plot.subtitle = element_text(face = "italic", size = 20),
plot.title = element_text(size = 20)
)
#ggsave(here("figures", "jo_sankey.jpg"), width=8, height = 5.5, dpi="retina", bg = "white", units="in")Table:
# Extract Probabilities
jo_prob_matrix <- as.matrix(jo_output$class_counts$transitionProbs$probability)
# Label Classes
c1_labels <- c("Pro-Math \nwith Elevated Utility Value \n(46%)",
"Math Ambivalent \nwith Minimal Utility Value\n(18%)",
"Math Ambivalent \nwith Elevated Utility Value\n(19%)",
"Anti-Math \nwith Minimal Utility Value\n(17%)")
c2_labels <- c("Pro-Science \nwith Elevated Utility Value\n(30%)",
"Science Ambivalent \nwith Minimal Utility Value\n(26%)",
"Science Ambivalent \nwith Elevated Utility Value\n(8%)",
"Anti-Science \nwith Minimal Utility Value\n(36%)")
# Number of Classes for each LCA
C1 <- length(c1_labels)
C2 <- length(c2_labels)
# Format Probability Table
jo_df <- matrix(jo_prob_matrix, nrow = C1, ncol = C2, byrow = FALSE)
rownames(jo_df) <- c1_labels
colnames(jo_df) <- c2_labels
t_matrix <- as.data.frame(jo_df) %>%
rownames_to_column(var = "Math Attitudes")
# Create Probability Table
t_matrix %>%
gt(rowname_col = "Math Attitudes") %>%
tab_stubhead(label = "Math Attitudes") %>%
tab_header(
title = md("**Joint Distribution Matrix**"),
subtitle = md("**Distribution Math Attitude Classes (Rows) conditioned on Science Attitude Classes (Columns)**")) %>%
fmt_number(2:4,decimals = 3) %>%
tab_spanner(label = "Science Attitudes",columns = 2:(C2+1))#%>% | Joint Distribution Matrix | ||||
| Distribution Math Attitude Classes (Rows) conditioned on Science Attitude Classes (Columns) | ||||
| Math Attitudes |
Science Attitudes
|
|||
|---|---|---|---|---|
| Pro-Science with Elevated Utility Value (30%) | Science Ambivalent with Minimal Utility Value (26%) | Science Ambivalent with Elevated Utility Value (8%) | Anti-Science with Minimal Utility Value (36%) | |
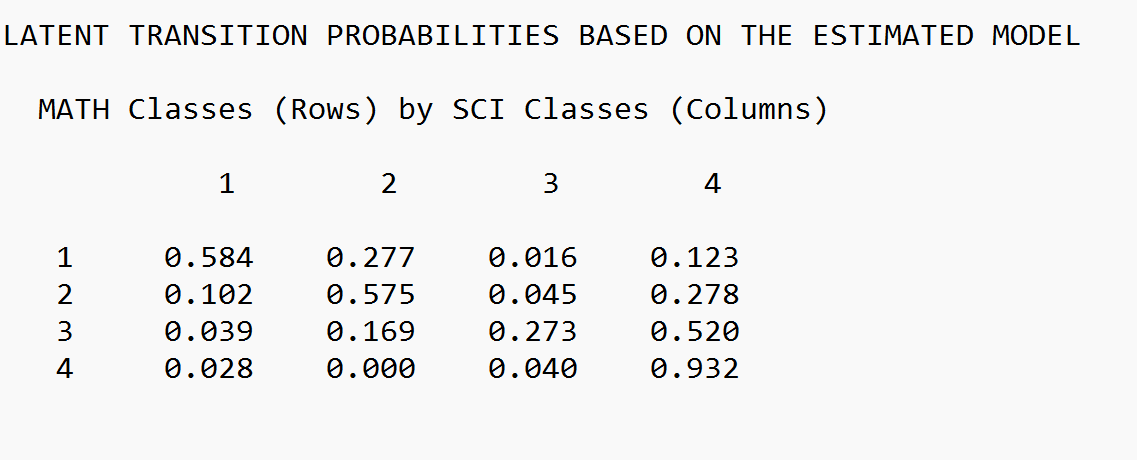
| Pro-Math with Elevated Utility Value (46%) | 0.584 | 0.277 | 0.016 | 0.123 |
| Math Ambivalent with Minimal Utility Value (18%) | 0.102 | 0.575 | 0.045 | 0.278 |
| Math Ambivalent with Elevated Utility Value (19%) | 0.039 | 0.169 | 0.273 | 0.520 |
| Anti-Math with Minimal Utility Value (17%) | 0.028 | 0.000 | 0.040 | 0.932 |
#gtsave("matrix.docx")Always check the output to make sure the table is correct!